setwd("C:/Users/Vicente/repo/ClimatePanama/ClimatePanamaBook")
library(dplyr)
library(ggplot2)
library(readxl)
library(tidyr)1 Gather STRI data
1.1 Libraries
The packages we utilize are dplyr(Wickham et al. 2023) and tidyr(Wickham, Vaughan, and Girlich 2024) for data wrangling, ggplot2(Wickham 2016) for data visualization and readxl(Wickham and Bryan 2023) for reading excel files.
1.2 Data
The stri_met_stations.csv contains links and metadata for all sites listed in https://biogeodb.stri.si.edu/physical_monitoring/
DIRTOMET<-"../tables/stri_met_stations.csv"
met_stations<- read.csv(DIRTOMET)
all_stations<-unique(met_stations$alias)
print(all_stations) [1] "CELESTINO" "AVA" "BCICLEAR" "BCIELECT" "BOCAS"
[6] "PCULEBRA" "FORTUNA" "GALETASTRI" "PNM" "SANBLAS"
[11] "SHERMAN" The following function calculates monthly precipitation given the standard format of the STRI data, by default it only includes values label as “good”
monthly_precip<- function(file, include=("good"),site_name){
sheets<-read.csv(file) %>% filter(chk_note %in% include)
if (nrow(sheets)<30){
stop("Error: not enough days with good measurments")}
daily_precip<-sheets%>%group_by(date)%>%summarize(daily_precip= sum(ra, na.rm = T))
monthly_precip <- daily_precip %>% mutate(month_year = as.Date(paste0(format(as.Date(date, "%d/%m/%Y"), "%Y-%m"), "-01")))%>%
group_by(month_year) %>% summarize(monthly_precip = sum(daily_precip, na.rm = T))
monthly_precip$site<-site_name
return(monthly_precip)
}The download_met function download the file , unzips it and then erases the zip file.
download_met<- function(site,csv){
link<-csv[csv$alias==site,'link']
destfile<-paste0("../data_ground/met_data/STRI_data/",csv[csv$alias==site,'alias'],".zip")
download.file(link,destfile =destfile , mode = "wb")
unzip(destfile, exdir = paste0("../data_ground/met_data/STRI_data/",csv[csv$alias==site,'alias']))
file.remove(destfile)
}1.3 Download data from sites with regular format
This sites only have a single csv file. Data sets provided by the Physical Monitoring Program of the Smithsonian Tropical Research Institute(Smithsonian Tropical Research Institute 2023).
sites <- c("PCULEBRA", "FORTUNA", "PNM", "SHERMAN", "AVA", "BCICLEAR", "BCIELECT", "BOCAS")
for (site in sites) {
site_dir <- paste0("../data_ground/met_data/STRI_data/", met_stations[met_stations$alias == site, 'alias'])
if (!dir.exists(site_dir)) {
download_met(site, met_stations)
}
files_in_site <- list.files(site_dir, recursive = TRUE, pattern = ".csv", full.names = TRUE)
This_site <- tryCatch(monthly_precip(files_in_site[1], include = c("good", "adjusted"), site_name = met_stations[met_stations$alias == site, 'alias']),
error = function(e) NULL)
if (!is.null(This_site)) {
write.csv(This_site, paste0(site_dir, '.csv'))
}
rm(This_site)
}1.4 Download data from sites with non-regular format
1.4.1 Galeta station special case
GALETASTRI and GALETALAB share the same values in the first years, then in 2002 the values are different GALETALAB stops in 2007.
We accept the adjusted values for GALETASTRI which is the tower station, the metadata inform us that the ACP close-by station Limon bay was used for the gap filling.
site= "GALETASTRI"
download_met(site, met_stations)[1] TRUEfiles_in_site<-list.files(paste0("../data_ground/met_data/STRI_data/",met_stations[met_stations$alias==site,'alias']),recursive = T, pattern = ".csv", full.names = T)
This_site<-monthly_precip(files_in_site[2],include=c("good","adjusted"), site_name = met_stations[met_stations$alias==site,'alias'])
if (exists('This_site')) {
write.csv(This_site, paste0('../data_ground/met_data/STRI_data/', met_stations[met_stations$alias == site, 'alias'], '.csv'))
}
remove(This_site)GALETALAB has a second sensor in the laboratory. It has no notes, and does not follow the format of the other stations
GALETALAB<- read.csv(files_in_site[1])%>%mutate(month_year = as.Date(paste0(format(as.Date(date.time..yyyy.mm.dd.hh.mm.ss., "%Y-%m-%d"), "%Y-%m"), "-01")))%>%
group_by(month_year)%>% summarise(monthly_precip = sum(ra.mm., na.rm = T))%>% mutate(site="GALETALAB")
if (exists('GALETALAB')) {
write.csv(GALETALAB, paste0('../data_ground/met_data/STRI_data/', "GALETALAB", '.csv'))
}
remove(GALETALAB)1.4.2 San Blas station special case
The san blas station does not follow the standard format and only provides with the summaries.
site= "SANBLAS"
download.file(met_stations[met_stations$alias==site,'link'],destfile =paste0("../data_ground/met_data/STRI_data/",met_stations[met_stations$alias==site,'alias'],".xlsx") , mode = "wb")
file<- read_excel(paste0("../data_ground/met_data/STRI_data/",met_stations[met_stations$alias==site,'alias'],".xlsx"), sheet = 4)
file<-file[2:9,1:13]
SANBLAS<- file%>%rename('01'='...2','02'= '...3','03'= '...4','04'= '...5','05'= '...6','06'= '...7','07'= '...8','08'= '...9','09'= '...10','10'= '...11','11'= '...12','12'= '...13')%>%
pivot_longer(cols = c('01','02','03','04','05','06','07','08','09','10','11','12'), names_to = 'month', values_to = 'monthly_precip')%>%mutate(month_year = as.Date(paste0(format(as.Date(paste0('01/',month,'/2000'), "%d/%m/%Y"), "%Y-%m"), "-01")))%>%
select(month_year, monthly_precip)%>%mutate(site="SANBLAS")
if (exists('SANBLAS')) {
write.csv(SANBLAS, paste0('../data_ground/met_data/STRI_data/', "SANBLAS", '.csv'))
}
remove(SANBLAS)
file.remove(paste0("../data_ground/met_data/STRI_data/",met_stations[met_stations$alias==site,'alias'],".xlsx"))[1] TRUE1.4.3 Agua salud station special case
This site has 2 sensors within meters of each other
site= "CELESTINO"
download_met(site, met_stations)[1] TRUEfiles_in_site<-list.files(paste0("../data_ground/met_data/STRI_data/",met_stations[met_stations$alias==site,'alias']),
recursive = T, pattern = ".csv", full.names = T)
This_siteN=monthly_precip(files_in_site[1],include=c("good","adjusted"), site_name = met_stations[met_stations$alias==site,'alias'])
This_siteS=monthly_precip(files_in_site[2],include=c("good","adjusted"), site_name = met_stations[met_stations$alias==site,'alias'])
plot(This_siteN$monthly_precip, This_siteS$monthly_precip)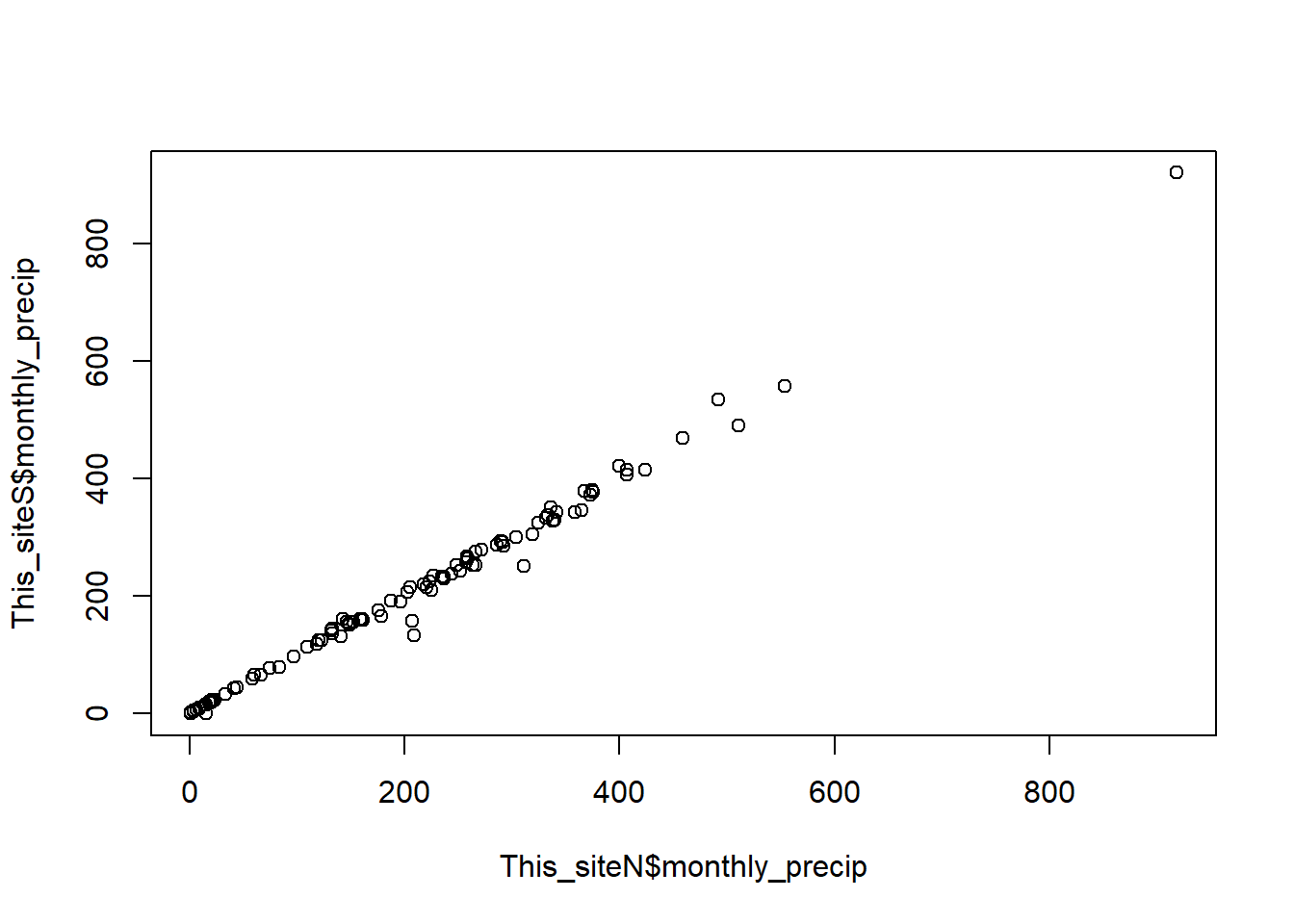
Merge both sensors and average them.
CELESTINO= merge(This_siteN, This_siteS, by = "month_year", all = TRUE)%>%mutate(monthly_precip = rowMeans(.[, c("monthly_precip.x", "monthly_precip.y")], na.rm = TRUE))%>%
select(month_year, monthly_precip)%>%mutate(site="CELESTINO")
if (exists('CELESTINO')) {
write.csv(CELESTINO, paste0('../data_ground/met_data/STRI_data/', met_stations[met_stations$alias == site, 'alias'], '.csv'))
}
remove(CELESTINO)1.5 Combine all the downloaded sites
all <- data.frame()
for (site in all_stations) {
thecsv <- file.path('../data_ground/met_data/STRI_data', paste0(site, '.csv'))
if (file.exists(thecsv)) {
append_stations <- read.csv(thecsv)
all <- rbind(all, append_stations)
}
}
all<- all%>%select(month_year, monthly_precip, site)%>%mutate(month_year = as.Date(month_year))Write the STRI_monthltPrecip file
write.csv(all, paste0('../data_ground/met_data/STRI_data/', "STRI_monthlyPrecip", '.csv'))