library(dplyr)
library(terra)
library(readxl)
library(kableExtra)
library(purrr)
library(sp)
library(tibble)
library(ggplot2)5 Rain Gauges vs Gridded Precipitation Analysis
5.1 Library
We utilize the terra package for rasters (Hijmans 2024). The package KableExtra is for table visualizations(Hao Zhu 2024). The sp package is used for calculating distances (Pebesma and Bivand 2005). Packages purr and tiblle are for data wrangling Müller and Wickham (2023)
Read in files for the analysis
# Read in the data
data_ground<- read.csv("../tables/precipitation.csv")
striStation<- read.csv("../tables/stri_met_stations.csv")
acpStation<- read.csv("../data_ground/met_data/ACP_data/cleanedVV/ACP_met_stations.csv")
#Global variables
extentMap <- terra::ext(c(-80.2,-79.4, 8.8, 9.5))5.2 Functions
calculate_climatology<- function(minyear,maxyear,data){
climatology<- data %>%
dplyr::select(year,site,annual,jfma,gap_fill)%>%
filter(year>=minyear&year<=maxyear)%>%
group_by(site)%>%
summarize(annual=mean(annual,na.rm=TRUE),
jfma=mean(jfma,na.rm=TRUE),
gapFill_percentage = mean(gap_fill, na.rm = TRUE) * 100)
return(climatology)
}
annual_pan<-function(list,extent){
b<-terra::rast(list)
c<-terra::app(b,sum)
d<-terra::crop(c,extent)
return(d)
}
jfma_pan <- function(list, extent) {
b <- terra::rast(head(list, 4))
c <- terra::app(b, sum)
d <- terra::crop(c, extent)
return(d)
}
find_nearest_non_na <- function(raster_object, coordinate) {
non_na_cells <- which(!is.na(terra::values(raster_object)))
non_na_coords <- xyFromCell(raster_object, non_na_cells)
dists <- sp::spDists(x = matrix(coordinate, nrow = 1), y = non_na_coords, longlat = TRUE)
nearest_cell <- non_na_cells[which.min(dists)]
return(terra::values(raster_object)[nearest_cell])
}
extract_land <- function(list_object, coordinate) {
result <- numeric(nrow(coordinate))
for (i in seq_len(nrow(coordinate))) {
extracted_value <- terra::extract(list_object, coordinate[i, ], na.rm = FALSE)[1,2]
result[i] <- ifelse(is.na(extracted_value), find_nearest_non_na(list_object, as.matrix(coordinate[i, ])), extracted_value)
}
return(result)
}
find_nearest_non_na_id <- function(raster_object, coordinate) {
non_na_cells <- which(!is.na(terra::values(raster_object)))
non_na_coords <- xyFromCell(raster_object, non_na_cells)
dists <- sp::spDists(x = matrix(coordinate, nrow = 1), y = non_na_coords, longlat = TRUE)
nearest_cell <- non_na_cells[which.min(dists)]
return(nearest_cell)
}
extract_row_column <- function(list_object, coordinate) {
result <- character(nrow(coordinate)) # Use character vector to store "row,col"
for (i in seq_len(nrow(coordinate))) {
extracted_value <- terra::extract(list_object, coordinate[i, ], na.rm = FALSE)[1, 2]
extracted_id <- terra::extract(list_object, coordinate[i, ], na.rm = FALSE,cells=TRUE)[1,3]
result[i] <- ifelse(is.na(extracted_value), find_nearest_non_na_id(list_object,as.matrix(coordinate[i,])), extracted_id)
}
return(result)
}
#stats calculation
stats<-function(predicted,observed){
correlation<-cor.test(observed,predicted)$estimate
rmse<-sqrt(mean((predicted-observed)^2 , na.rm = TRUE ) )
bias<- mean((predicted-observed),na.rm=TRUE)
mae<- mean(abs(predicted-observed), na.rm = TRUE)
done<-c(correlation,rmse,bias,mae)
return(done)
}List of data sets, their temporal extents and the location of the climatology tiffs for each of them
datasets_info <- list(
list(start_year = 1979, end_year = 2013, dataset_name = "CHELSA 1.2",column_name="CHELSA1.2", temporal = "1979-2013",path = "../data_reanalysis/CHELSA 1.2/"),
list(start_year = 1981, end_year = 2010, dataset_name = "CHELSA 2.1",column_name="CHELSA2.1", temporal = "1981-2010", path = "../data_reanalysis/CHELSA 2.1/"),
list(start_year = 2003, end_year = 2016, dataset_name = "CHELSA EarthEnv",column_name="CHELSA_EarthEnv", temporal = "2003-2016",path = "../data_reanalysis/CHELSA EarthEnv/climatology"),
list(start_year = 1980, end_year = 2009, dataset_name = "CHPclim",column_name="CHPclim", temporal = "1980-2009", path = "../data_reanalysis/CHPclim/"),
list(start_year = 1970, end_year = 2000, dataset_name = "WorldClim 2.1",column_name="WorldClim2.1", temporal = "1970-2000", path = "../data_reanalysis/WorldClim 2.1"),
list(start_year = 1981, end_year = 2016, dataset_name = "CHIRPS 2.0",column_name="CHIRPS2.0",temporal = "1981-2016", path = "../data_reanalysis/CHIRPS 2.0/climatology"),
list(start_year = 1979, end_year = 2016, dataset_name = "CHELSA-W5E5v1.0",column_name="CHELSAW5E5v1.0", temporal = "1979-2016",path = "../data_reanalysis/CHELSA-W5E5v1.0/climatology"),
list(start_year = 1981, end_year = 2010, dataset_name = "TerraClimate",column_name="TerraClimate", temporal = "1981-2010", path = "../data_reanalysis/TerraClimate"),
list(start_year = 1979, end_year = 2013, dataset_name = "PBCOR CHELSA 1.2",column_name="PBCOR_CHELSA1.2", temporal = "1979-2013", path = "../data_reanalysis/PBCOR/CHELSA 1.2 corrected"),
list(start_year = 1980, end_year = 2009, dataset_name = "PBCOR CHPclim",column_name="PBCOR_CHPclim", temporal = "1980-2009", path = "../data_reanalysis/PBCOR/CHPclim corrected"),
list(start_year = 1970, end_year = 2000, dataset_name = "PBCOR WorldClim 2.1",column_name="PBCOR_WorldClim2.1", temporal = "1970-2000", path = "../data_reanalysis/PBCOR/WorldClim corrected")
)5.3 Climatologies
Loop through each dataset and calculate climatology
climatologies <- datasets_info %>%
map_df(function(info) {
calculate_climatology(info$start_year, info$end_year, data_ground) %>%
mutate(source = info$dataset_name)}) %>%
left_join(acpStation[, c('site', 'latitude', 'longitude', 'Elevation')], by = c("site" = "site")) %>%
left_join(striStation[, c('alias', 'latitude', 'longitude')], by = c("site" = "alias")) %>%
mutate(latitude = ifelse(is.na(latitude.x), latitude.y, latitude.x),
longitude = ifelse(is.na(longitude.x), longitude.y, longitude.x),
longitude = -1 * abs(longitude)) %>%
dplyr::select(site, latitude, longitude, source, annual, jfma, Elevation, gapFill_percentage)
#extract the values at the coordinates for climatologies
for (source in datasets_info) {
files <- list.files(source$path, pattern = "\\.tif$", full.names = TRUE)
annual_data <- annual_pan(files, extentMap)
climatologies[[source$dataset_name]] <- extract_land(annual_data,climatologies[,3:2])
climatologies[[paste0(source$dataset_name,"_cell")]]<-extract_row_column(annual_data,climatologies[,3:2])
jfma_data = jfma_pan(files, extentMap)
column_name = paste(source$dataset_name, "_jfma", sep = "")
climatologies[[column_name]] = extract_land(jfma_data, climatologies[,3:2])
}
write.csv(climatologies,file="../tables/climatologies.csv")#Briefly analyse the gap filling effect on site and Gridded dataset
ggplot(climatologies, aes(x = site, y = gapFill_percentage)) +
geom_boxplot() +
labs(title = "Gap Fill Percentage by Site",
x = "Site",
y = "Gap Fill Percentage (%)") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1))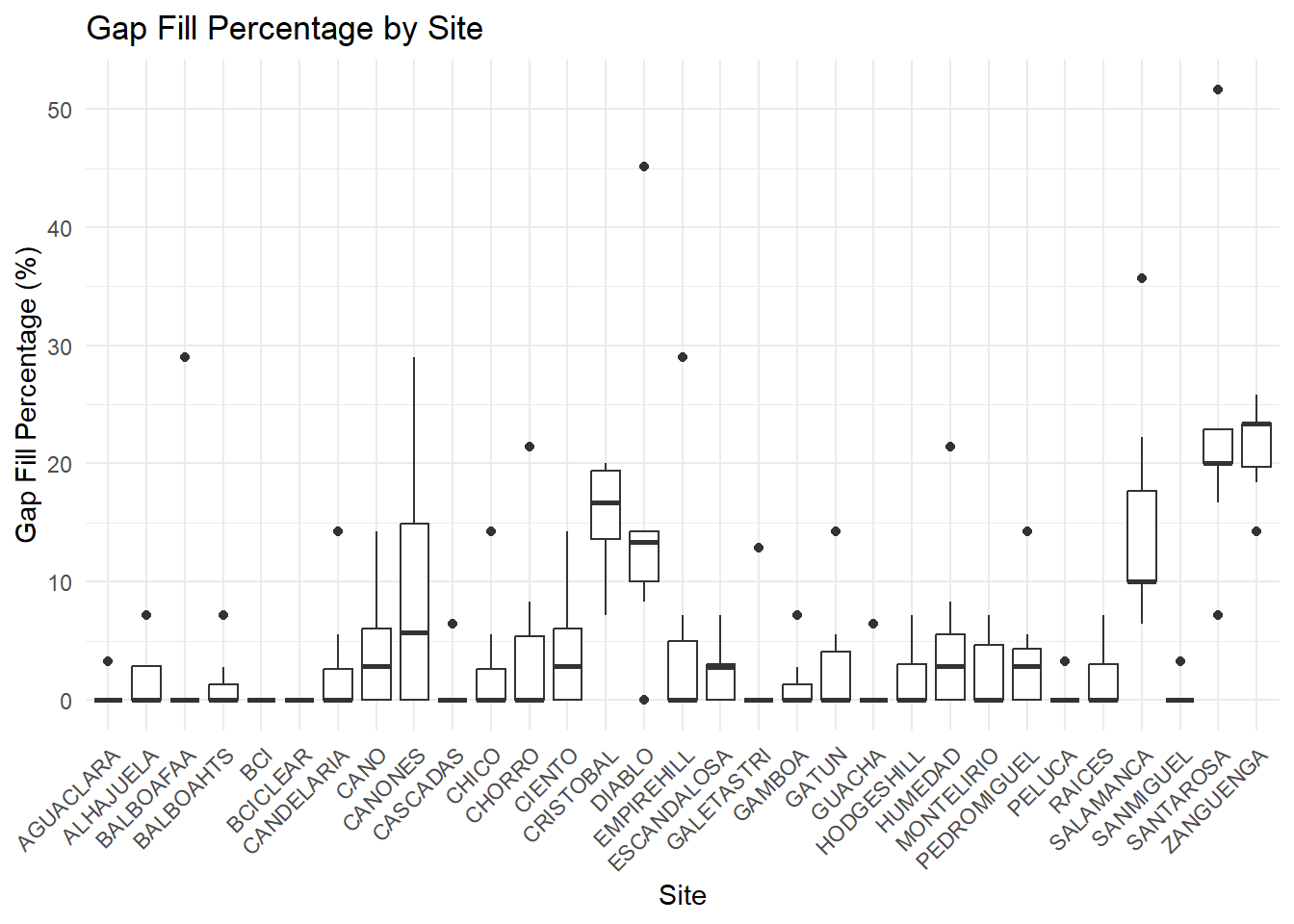
ggplot(climatologies, aes(x = source, y = gapFill_percentage)) +
geom_boxplot() +
labs(title = "Gap Fill Percentage by Site",
x = "Source",
y = "Gap Fill Percentage (%)") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1))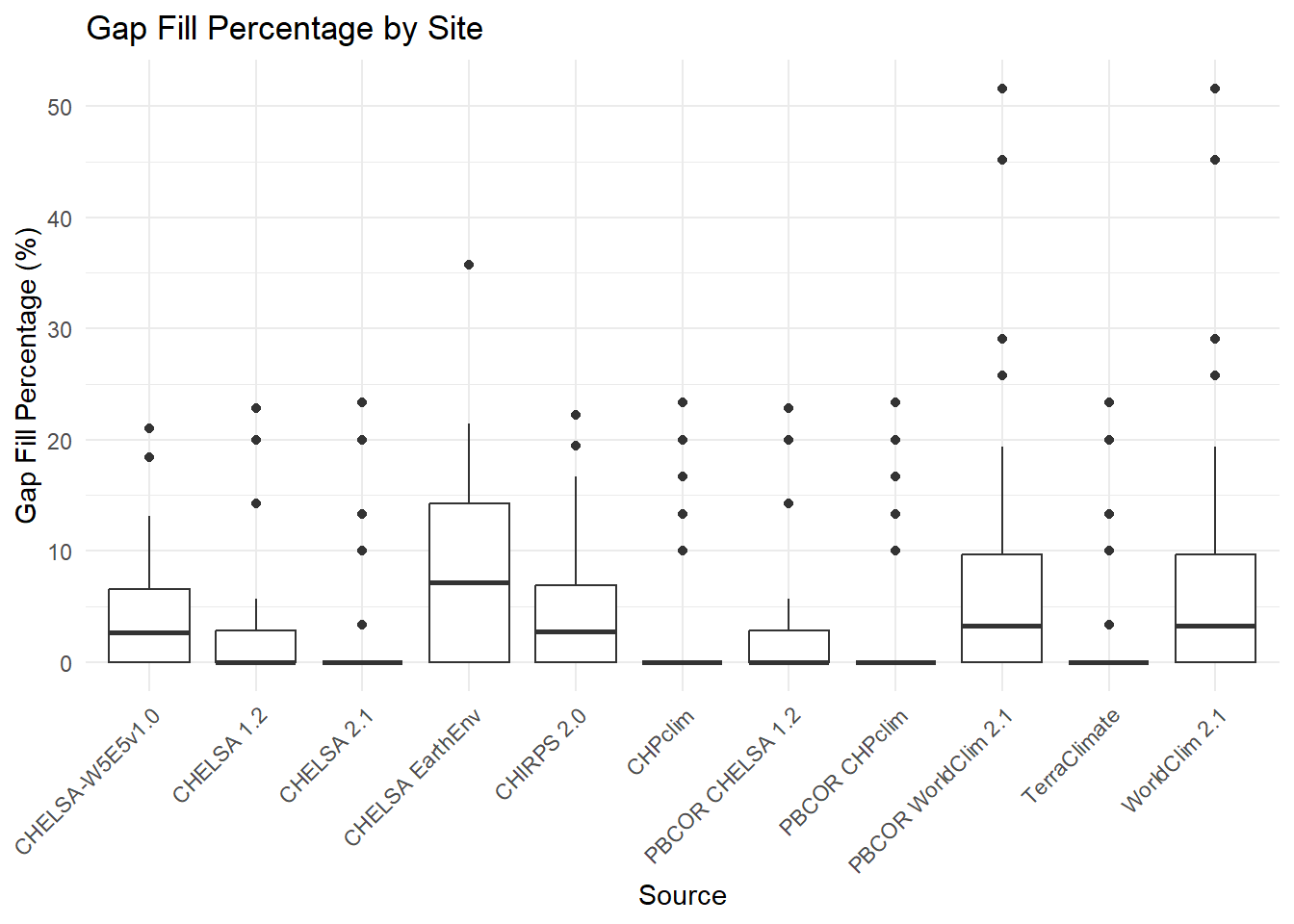
sources_unique <- unique(climatologies$source)
df<- data.frame()
for (source in sources_unique) {
data_column <- paste0(source)
unique_values <- nrow(unique(climatologies[climatologies$source == source, paste0(source,"_cell")]))
# Find sites with the same value
sites_with_same_value <- climatologies[climatologies$source == source, ] %>%
group_by(.data[[paste0(source, "_cell")]]) %>%
filter(n() > 1) %>%
summarize(sites = paste(site, collapse = ", "))
cat(paste("Source:", source, "\n"))
cat(paste(" - Unique raster cell:", unique_values, "\n"))
cat(paste(" - Sites in the same cell:", sites_with_same_value$sites, "\n"))
}Source: CHELSA 1.2
- Unique raster cell: 30
- Sites in the same cell: BCI, BCICLEAR
Source: CHELSA 2.1
- Unique raster cell: 30
- Sites in the same cell: BCI, BCICLEAR
Source: CHELSA EarthEnv
- Unique raster cell: 30
- Sites in the same cell: BCI, BCICLEAR
Source: CHPclim
- Unique raster cell: 28
- Sites in the same cell: BCI, BCICLEAR
- Sites in the same cell: CASCADAS, EMPIREHILL
- Sites in the same cell: BALBOAHTS, DIABLO
Source: WorldClim 2.1
- Unique raster cell: 30
- Sites in the same cell: BCI, BCICLEAR
Source: CHIRPS 2.0
- Unique raster cell: 28
- Sites in the same cell: BCI, BCICLEAR
- Sites in the same cell: CASCADAS, EMPIREHILL
- Sites in the same cell: BALBOAHTS, DIABLO
Source: CHELSA-W5E5v1.0
- Unique raster cell: 30
- Sites in the same cell: BCI, BCICLEAR
Source: TerraClimate
- Unique raster cell: 27
- Sites in the same cell: BCI, BCICLEAR
- Sites in the same cell: EMPIREHILL, HODGESHILL
- Sites in the same cell: BALBOAFAA, BALBOAHTS, DIABLO
Source: PBCOR CHELSA 1.2
- Unique raster cell: 27
- Sites in the same cell: BCI, BCICLEAR
- Sites in the same cell: CASCADAS, EMPIREHILL
- Sites in the same cell: BALBOAHTS, DIABLO
- Sites in the same cell: CRISTOBAL, GALETASTRI
Source: PBCOR CHPclim
- Unique raster cell: 28
- Sites in the same cell: BCI, BCICLEAR
- Sites in the same cell: CASCADAS, EMPIREHILL
- Sites in the same cell: BALBOAHTS, DIABLO
Source: PBCOR WorldClim 2.1
- Unique raster cell: 27
- Sites in the same cell: BCI, BCICLEAR
- Sites in the same cell: CASCADAS, EMPIREHILL
- Sites in the same cell: BALBOAHTS, DIABLO
- Sites in the same cell: CRISTOBAL, GALETASTRI Compute a results data frame
results <- data.frame(
Source = character(),
Correlation = numeric(),
RMSE = numeric(),
Bias = numeric(),
MAE = numeric(),
stringsAsFactors = FALSE
)
results_jfma <-results
for (i in 1:length(datasets_info)) {
source <- datasets_info[[i]]$dataset_name
predicted <- climatologies[climatologies$source == source,][[source]]
observed <- climatologies[climatologies$source == source,][["annual"]]
stats_result <- stats(predicted, observed)
results <- rbind(results, data.frame(
Source = source,
Correlation = stats_result[1],
RMSE = stats_result[2],
MAE = stats_result[4],
Bias = stats_result[3],
stringsAsFactors = FALSE
))
}
for (i in 1:length(datasets_info)) {
source <- datasets_info[[i]]$dataset_name
column=paste0(source,"_jfma")
predicted <- climatologies[climatologies$source == source,][[column]]
observed <- climatologies[climatologies$source == source,][["jfma"]]
stats_result <- stats(predicted, observed)
results_jfma <- rbind(results_jfma, data.frame(
Source = source,
Correlation = stats_result[1],
RMSE = stats_result[2],
MAE = stats_result[4],
Bias = stats_result[3],
stringsAsFactors = FALSE
))
}
results$Source[results$Source == "CHIRPS 2.0"] <- "CHIRPS v2.0"
results$Source[results$Source == "CHPclim"] <- "CHPclim v1.0"
results_jfma$Source[results_jfma$Source == "CHIRPS 2.0"] <- "CHIRPS v2.0"
results_jfma$Source[results_jfma$Source == "CHPclim"] <- "CHPclim v1.0"
order_vec <- c("CHELSA 1.2","CHELSA 2.1","CHELSA EarthEnv","CHELSA-W5E5v1.0","CHIRPS v2.0","CHPclim v1.0",
"TerraClimate","WorldClim 2.1","PBCOR CHELSA 1.2","PBCOR CHPclim","PBCOR WorldClim 2.1")
results <- results[match(order_vec, results$Source), ]
results_jfma <- results_jfma[match(order_vec, results_jfma$Source), ]
#save both results as csv in tables folder
write.csv(results, file= "../tables/spatial_variation_annual.csv")
write.csv(results_jfma, file= "../tables/spatial_variation_jfma.csv")Tables for visualizations
annual_table<-results %>%mutate(Correlation = round(Correlation, 2), RMSE= round(RMSE,0),Bias=round(Bias,0),MAE=round(MAE,0)) %>%
rownames_to_column(var = "rowname") %>%
dplyr::select(-rowname) %>%
rename("Gridded Climate Product" = "Source") %>% rename("Mean bias (mm)"="Bias")%>%
kbl(caption = "Spatial variation among 31 sites in total annual precipitation") %>%
kable_classic(full_width = F)
annual_table%>%save_kable("../plots/spatial_annual_variation.png",density=900)
annual_table| Gridded Climate Product | Correlation | RMSE | MAE | Mean bias (mm) |
|---|---|---|---|---|
| CHELSA 1.2 | 0.82 | 310 | 250 | 53 |
| CHELSA 2.1 | 0.87 | 475 | 438 | 390 |
| CHELSA EarthEnv | 0.85 | 318 | 258 | 68 |
| CHELSA-W5E5v1.0 | 0.36 | 497 | 401 | -19 |
| CHIRPS v2.0 | 0.88 | 273 | 222 | 92 |
| CHPclim v1.0 | 0.86 | 371 | 326 | 245 |
| TerraClimate | 0.78 | 412 | 346 | -50 |
| WorldClim 2.1 | 0.65 | 402 | 330 | -24 |
| PBCOR CHELSA 1.2 | 0.75 | 377 | 306 | 107 |
| PBCOR CHPclim | 0.77 | 547 | 463 | 413 |
| PBCOR WorldClim 2.1 | 0.56 | 423 | 337 | -21 |
jfma_table<-results_jfma %>%mutate(Correlation = round(Correlation, 2), RMSE= round(RMSE,0),Bias=round(Bias,0),MAE=round(MAE,0)) %>%
rownames_to_column(var = "rowname") %>%
dplyr::select(-rowname) %>%
rename("Gridded Climate Product" = "Source") %>% rename("Mean bias (mm)"="Bias")%>%
kbl(caption = "Spatial variation among 31 sites in January-to-April precipitation") %>%
kable_classic(full_width = F)
jfma_table%>%save_kable("../plots/spatial_jfma_variation.png",density=900)
jfma_table| Gridded Climate Product | Correlation | RMSE | MAE | Mean bias (mm) |
|---|---|---|---|---|
| CHELSA 1.2 | 0.63 | 107 | 63 | -35 |
| CHELSA 2.1 | 0.61 | 105 | 69 | -6 |
| CHELSA EarthEnv | 0.66 | 91 | 61 | 4 |
| CHELSA-W5E5v1.0 | 0.15 | 125 | 83 | -10 |
| CHIRPS v2.0 | 0.82 | 81 | 49 | -22 |
| CHPclim v1.0 | 0.86 | 91 | 57 | -46 |
| TerraClimate | 0.84 | 112 | 74 | -64 |
| WorldClim 2.1 | 0.47 | 121 | 84 | -52 |
| PBCOR CHELSA 1.2 | 0.60 | 108 | 66 | -32 |
| PBCOR CHPclim | 0.81 | 89 | 54 | -32 |
| PBCOR WorldClim 2.1 | 0.32 | 131 | 93 | -57 |
5.4 Seasonal variation(12-month)
month <- read.csv("../tables/monthly_ground.csv")
sites <- read.csv("../tables/dfprecip.csv")
#Filter data based on conditions
acp38 <- month %>%
filter(site %in% c("AGUACLARA", "SANMIGUEL", "BCI", "PELUCA", "BCICLEAR", "GUACHA", "CASCADAS", "PEDROMIGUEL", "GAMBOA") &
Year >= 1970 & Year <= 2016) %>%
mutate(monthlyPrecip = as.numeric(monthlyPrecip))
acp38_full_extent <- acp38 %>%
group_by(site, Month) %>%
summarise(mon_mean = mean(monthlyPrecip, na.rm = TRUE)) %>%
mutate(temporal = "1970-2016", dataset_name = "Ground")
# Calculate climatology of the ground data for every specific temporal extent
acp38_ag <- lapply(datasets_info, function(dataset) {
acp38 %>%
filter(Year >= dataset$start_year & Year <= dataset$end_year) %>%
group_by(site, Month) %>%
summarise(mon_mean = mean(monthlyPrecip, na.rm = TRUE)) %>%
mutate(temporal = dataset$temporal, dataset_name = dataset$dataset_name)
}) %>% bind_rows()
# Combine the two data frames
acp38_ag <- bind_rows(acp38_ag, acp38_full_extent)
#calculate climatology of the ground data for every specific temporal extent
# Match lat and long
acp38_ag$long_dd <- sites$long_dd[match(acp38_ag$site, sites$site)]
acp38_ag$lat_dd <- sites$lat_dd[match(acp38_ag$site, sites$site)]
## loop for extraction of values of raster reanalyis climatologies for specific sites and month.
for (i in 1:length(datasets_info)) {
files_list <- list.files(datasets_info[[i]]$path, pattern = "\\.tif$", full.names = TRUE)
raster_stack <- terra::rast(files_list)
acp38_ag[[datasets_info[[i]]$column_name]] <- NA
for (j in 1:12) {
acp38_ag[acp38_ag$Month == j, datasets_info[[i]]$column_name] <- extract_land(raster_stack[[j]], acp38_ag[acp38_ag$Month == j, c("long_dd", "lat_dd")])
}
}
write.csv(acp38_ag,"../tables/acp38_ag.csv")After extracting values, we can run some statistics
sites_unique <- unique(acp38_ag$site)
all_correlations <- data.frame( site = character(),
dataset=character(),
Correlation = numeric(),
RMSE = numeric(),
stringsAsFactors = FALSE)
#loop over the dataset and each of the sites for calculating the correlation and the rmse
for (j in seq_along(datasets_info)){
print(datasets_info[[j]]$dataset_name)
for (i in seq_along(sites_unique)) {
temporal_range <- datasets_info[[j]]$temporal
name <- datasets_info[[j]]$dataset_name
col_name <- datasets_info[[j]]$column_name
site_data <- acp38_ag[acp38_ag$site == sites_unique[i] & acp38_ag$temporal == temporal_range & acp38_ag$dataset_name==name, ]
cor_test_result <- cor.test(site_data$mon_mean, unlist(site_data[,col_name]))$estimate
rmse<-sqrt(mean(unlist(site_data[,col_name])-site_data$mon_mean)^2)
new_row <- data.frame(site = sites_unique[i], dataset = name, Correlation = cor_test_result, RMSE = rmse)
all_correlations <- rbind(all_correlations, new_row)
}
}[1] "CHELSA 1.2"
[1] "CHELSA 2.1"
[1] "CHELSA EarthEnv"
[1] "CHPclim"
[1] "WorldClim 2.1"
[1] "CHIRPS 2.0"
[1] "CHELSA-W5E5v1.0"
[1] "TerraClimate"
[1] "PBCOR CHELSA 1.2"
[1] "PBCOR CHPclim"
[1] "PBCOR WorldClim 2.1"all_averages<- data.frame(datset=character(),
Correlation= numeric(),
RMSE=numeric())
datasets<- unique(all_correlations$dataset)
for (i in seq_along(datasets)){
dataset_name=datasets[i]
mean_correlation= mean(all_correlations[all_correlations$dataset==dataset_name,]$Correlation)
mean_rsme= mean(all_correlations[all_correlations$dataset==dataset_name,]$RMSE)
new_row= data.frame(dataset=dataset_name,Correlation=mean_correlation,RMSE=mean_rsme)
all_averages= rbind(all_averages,new_row)
}
write.csv(all_averages, file= "../tables/seasonal_variation.csv")Plot the statistics in a table
seasonality_table<-all_averages%>%mutate(Correlation = round(Correlation, 2), RMSE= round(RMSE,0)) %>%
rownames_to_column(var = "rowname") %>%
dplyr::select(-rowname) %>%
rename("Gridded Climate Product" = "dataset")%>%
kbl(caption = "Seasonal variation within 9 sites") %>%
kable_classic(full_width = F)
seasonality_table%>%save_kable("../plots/seasonal_variation.png",density=900)
seasonality_table| Gridded Climate Product | Correlation | RMSE |
|---|---|---|
| CHELSA 1.2 | 0.97 | 31 |
| CHELSA 2.1 | 0.97 | 42 |
| CHELSA EarthEnv | 0.94 | 24 |
| CHPclim | 0.96 | 29 |
| WorldClim 2.1 | 0.95 | 34 |
| CHIRPS 2.0 | 0.98 | 23 |
| CHELSA-W5E5v1.0 | 0.97 | 42 |
| TerraClimate | 0.93 | 33 |
| PBCOR CHELSA 1.2 | 0.97 | 28 |
| PBCOR CHPclim | 0.96 | 36 |
| PBCOR WorldClim 2.1 | 0.96 | 36 |
5.5 Interannual variability
# create a list which contains the temporal extent and location of files
datasets_yearly <- list(
list(dataset_name = "CHELSA 1.2",column_name="CHELSA1.2", temporal = "1979-2013",path = "../data_reanalysis/CHELSA 1.2/yearly"),
list(dataset_name = "CHELSA 2.1",column_name="CHELSA2.1", temporal = "1981-2010", path = "../data_reanalysis/CHELSA 2.1/yearly"),
list(dataset_name = "CHELSA EarthEnv",column_name="CHELSA_EarthEnv", temporal = "2003-2016",path = "../data_reanalysis/CHELSA EarthEnv/yearly"),
list(dataset_name = "CHIRPS 2.0",column_name="CHIRPS2.0",temporal = "1981-2016", path = "../data_reanalysis/CHIRPS 2.0/yearly"),
list(dataset_name = "CHELSA-W5E5v1.0",column_name="CHELSAW5E5v1.0", temporal = "1979-2016",path = "../data_reanalysis/CHELSA-W5E5v1.0/yearly"),
list(dataset_name = "TerraClimate",column_name="TerraClimate", temporal = "1979-2016", path = "../data_reanalysis/TerraClimate/yearly")
)Group by year and extract for each spatial point
acp38_year <- lapply(datasets_yearly, function(dataset) {
acp38 %>% group_by(Year,site)%>%
summarise(annual_precip=sum(monthlyPrecip))%>%
mutate(long_dd=sites$long_dd[match(site, sites$site)])%>%
mutate(lat_dd=sites$lat_dd[match(site,sites$site)])%>%
mutate(temporal = dataset$temporal, dataset_name = dataset$dataset_name)
})%>% bind_rows()
for (j in seq_along(datasets_yearly)){
print(datasets_yearly[[j]]$dataset_name)
files_list <- list.files(datasets_yearly[[j]]$path, pattern = ".tif", full.names = TRUE)
raster_stack<- terra::rast(files_list)
acp38_year[[datasets_yearly[[j]]$column_name]] <- NA
for (i in unique(acp38_year$Year)) {
if (any(grepl(i,names(raster_stack)))) {
layer_index<-grep(i, names(raster_stack))
raster_layer <- raster_stack[[layer_index]]
acp38_year[acp38_year$Year==as.integer(i),datasets_yearly[[j]]$column_name]<- extract_land(raster_layer,acp38_year[acp38_year$Year==as.integer(i),c("long_dd","lat_dd")])
} else {
}
}
}Calculate the statistics for interannual variability per site and compute averages across 9 sites
sites_unique_year <- unique(acp38_year$site)
all_correlations_year <- data.frame( site = character(),
dataset=character(),
Correlation = numeric(),
RMSE = numeric(),
stringsAsFactors = FALSE)
for (j in seq_along(datasets_yearly)){
print(datasets_yearly[[j]]$dataset_name)
for (i in seq_along(sites_unique_year)) {
temporal_range <- datasets_yearly[[j]]$temporal
name <- datasets_yearly[[j]]$dataset_name
col_name <- datasets_yearly[[j]]$column_name
site_data <- acp38_year[acp38_year$site == sites_unique_year[i] & acp38_year$temporal == temporal_range & acp38_year$dataset_name==name,]
cor_test_result <- cor.test(as.numeric(site_data$annual_precip), as.numeric(unlist(site_data[, col_name])), na.rm = TRUE)$estimate
rmse <- sqrt(mean((as.numeric(unlist(site_data[, col_name])) - site_data$annual_precip)^2, na.rm = TRUE))
new_row <- data.frame(site = sites_unique_year[i], dataset = name, Correlation = cor_test_result, RMSE = rmse)
all_correlations_year <- rbind(all_correlations_year, new_row)
}
}
summary_table <- all_correlations_year %>%
group_by(dataset) %>%
summarise(Correlation = mean(Correlation, na.rm = TRUE),
RMSE = mean(RMSE, na.rm = TRUE))
interannual_table <- summary_table %>%
mutate(Correlation = round(Correlation, 2), RMSE = round(RMSE, 0)) %>%
rename("Mean Correlation" = Correlation, "Mean RMSE" = RMSE) %>%
rownames_to_column(var = "Dataset") %>%
kbl(caption = "Summary of Correlation and RMSE by Dataset")%>% kable_classic(full_width = FALSE)
interannual_table%>%save_kable("../plots/interannual_table.png",density=900)
write.csv(summary_table, file= "../tables/interannual_variation.csv")
write.csv(acp38_year, file = "../tables/acp38_year.csv")
interannual_table